Introduction to GlucoKinaseDB
GlucoKinaseDB: A Centralized Resource for Diabetes Drug Target Information
Glucokinase (GK) is an essential enzyme that functions as a glucose sensor in the liver and pancreas, playing a crucial role in regulating blood sugar levels. This significant role makes GK an important target for the development of new drugs, particularly for type 2 diabetes.
Molecules that can influence the activity of glucokinase (GK), such as glucokinase activators (GKAs), are particularly significant. However, researchers face a major challenge: crucial information about these GK modulators (including activators, inhibitors, and compounds that disrupt GK’s interaction with its regulatory protein, GKRP) is dispersed across various scientific literature, patents, and chemical databases.
To address this issue, our research team initiated the development of a specialized, consolidated resource. The result is GlucoKinaseDB (GKDB), a comprehensive and manually curated database dedicated to glucokinase modulators.
What GKDB Offers
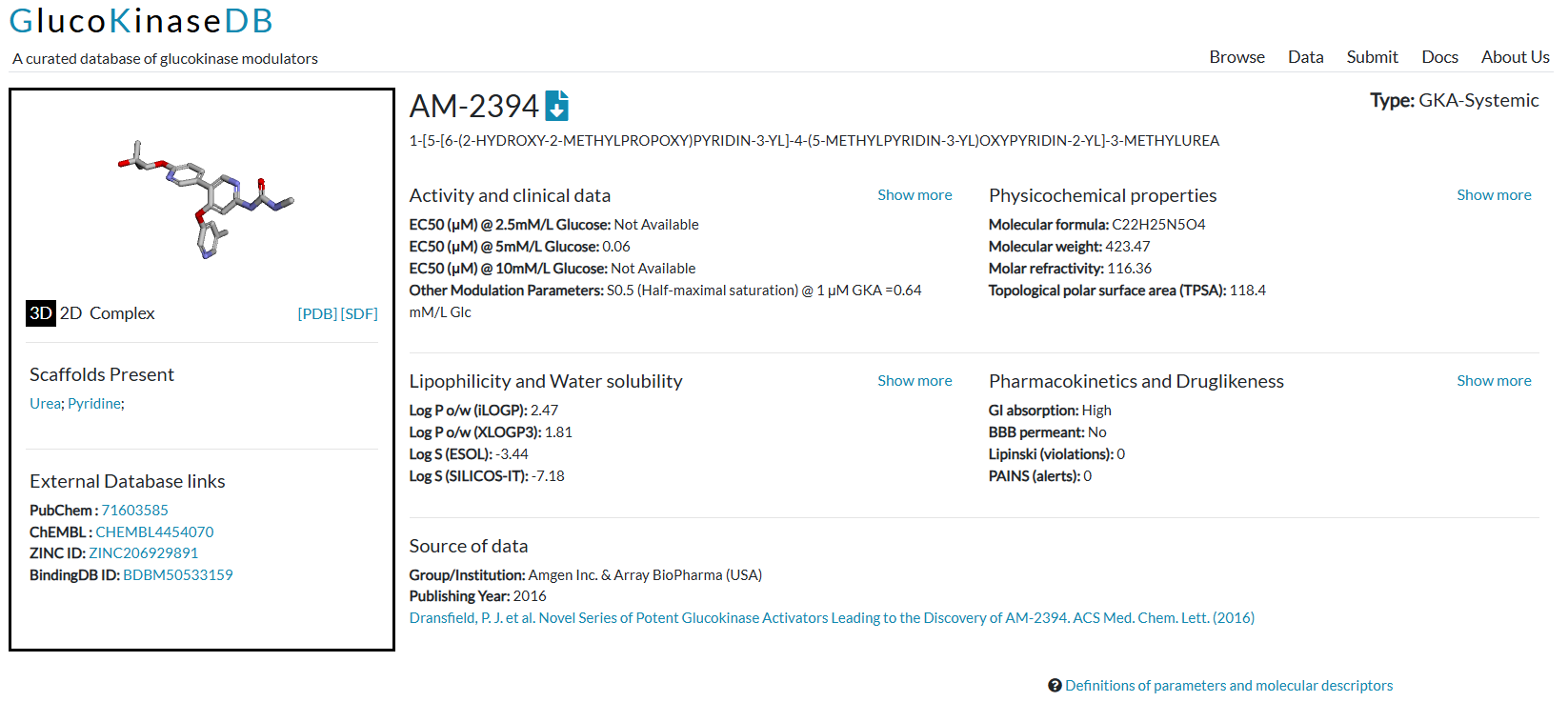
Centralized, Curated Data: GKDB houses structural and bioactivity information for over 1700 experimentally validated GK modulators, including GKAs, GK inhibitors (GKIs), and GK-GKRP disruptors. This data is meticulously gathered from publications, patents, and chemical repositories.
User-Friendly Interface & Tools: We designed GKDB with the researcher in mind:
Advanced search options for finding specific compounds. Interactive tables for easy data sorting and viewing.
In-browser 3D visualization of molecular structures.
Downloadable data (structures, bioactivity, descriptors) in multiple formats.
Direct links to original references and major external databases (e.g., PubChem, ChEMBL).
An API for computational access to the data.
Broadening the Scope: Applications of GKDB
While GKDB’s primary aim is to accelerate research on GK-targeted diabetes therapies, its curated dataset supports a wider range of applications:
Drug Discovery & Design: Aids in developing pharmacophore models and QSAR/QSPR studies to understand structure-activity relationships.
Computational Biology & Machine Learning: Provides high-quality, structured data suitable for training predictive models related to drug efficacy or properties.
Drug Repurposing: Consolidating data on known modulators can help identify compounds with potential for new therapeutic uses.
Targeting Specific Mechanisms: The inclusion of GK-GKRP disruptor data supports research into more targeted therapies, potentially hepatocyte-specific activation to avoid side effects like hypoglycemia associated with some GKAs.
Experimental Research Support: Provides easy access to information on compounds like GKAs used in laboratory studies investigating glucose sensing and hormone release.
Explore GKDB
Visit the database: https://glucokinasedb.in/
Access data via the API: https://glucokinasedb.in/dataset
Read our publication: Yadav, Bharti, Mathur (2023). Comp Biol Chem. https://doi.org/10.1016/j.compbiolchem.2023.107818
Our Collaborative Effort & My Role
Creating GKDB was a collaborative project, integrating expertise in biochemistry, data curation, and software development. While the team curated the data and provided biological insights, my specific contribution focused on the technical development. I designed and built the web platform using PHP, HTML, CSS, and JavaScript, set up and managed the MySQL database, and integrated essential interactive tools like 3Dmol.js for visualization and DataTables for structuring information. A key feature I implemented was the PHP-based API, enabling programmatic access for researchers using tools like Python or R.
We believe GKDB serves as a valuable hub for the scientific community investigating glucokinase, fostering advancements in both basic research and therapeutic development.
References
Moede, T., Leibiger, B., Vaca Sanchez, P. et al. Glucokinase intrinsically regulates glucose sensing and glucagon secretion in pancreatic alpha cells. Sci Rep 10, 20145 (2020). https://doi.org/10.1038/s41598-020-76863-z
Choi JM, Seo MH, Kyeong HH, Kim E, Kim HS. Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proc Natl Acad Sci U S A. 2013;110(25):10171-10176. doi:10.1073/pnas.1300457110
This post was drafted with assistance from various AI models to help share my project work more effectively. Please feel free to reach out if you spot any typos or have corrections!