Introduction to PepEngine
PepEngine: a database of synthetic peptides with the non-standard amino acids
Synthetic peptides are incredibly versatile molecules, playing vital roles in areas ranging from drug development to materials science. Some of the most interesting synthetic peptides incorporate non-standard amino acids, like α,β-Dehydrophenylalanine (ΔPhe or ΔF) and α-Aminoisobutyric acid (Aib). These unusual building blocks are known to significantly influence peptide structure, often inducing stable helical conformations or specific turns. Understanding these 3D structures is crucial for designing new peptides with desired functions, but finding consolidated structural information for peptides containing ΔF and Aib can be challenging.
To address this, our team developed PepEngine, a specialized, manually curated database focused exclusively on the structural details of synthetic peptides containing ΔF and Aib.
What PepEngine Provides
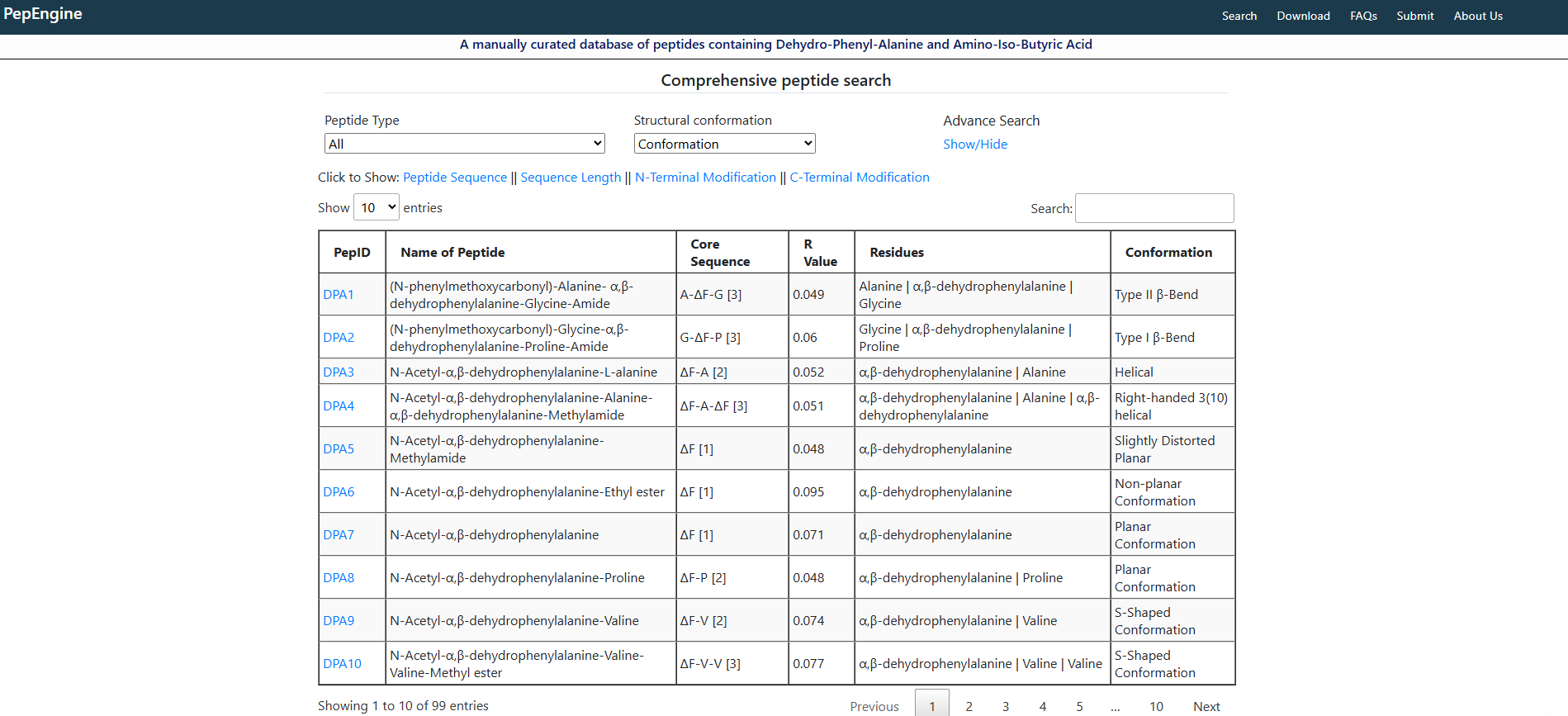
Focused Structural Data: Contains detailed structural information (including 3D coordinates) for synthetic peptides incorporating ΔF and/or Aib.
Manual Curation: Data is carefully gathered and verified from scientific literature, ensuring quality and relevance.
Interactive Exploration: In-browser 3D visualization of peptide structures using 3Dmol.js.
Organized data presentation using interactive tables (DataTables).
Easy access to source publication details.
Downloadable structural files (e.g., PDB format) for offline analysis and modeling.
Explore the Structures
Visit the PepEngine database: https://pepengine.in/
Read our publication: Yadav, Bharti, Srivastava & Mathur (2022). Int J Pept Res Ther. https://doi.org/10.1007/s10989-022-10362-9
We hope PepEngine facilitates new discoveries and designs in the fascinating world of synthetic peptides!
Our Collaborative Effort & My Role
PepEngine emerged from a collaborative effort combining expertise in peptide chemistry, structural biology, and bioinformatics. While the team focused on identifying relevant peptides and curating the structural data from literature, my role centered on the technical implementation of the database and web interface. I developed the platform using HTML, PHP, CSS, and JavaScript, managed the underlying MySQL database, and integrated the key visualization components like 3Dmol.js and the interactive DataTable library to ensure the structural data was easily accessible and explorable.
This post was drafted with assistance from various AI models to help share my project work more effectively. Please feel free to reach out if you spot any typos or have corrections!