Introduction to SEAS
Understanding Your Sample Subsets: An Introduction to SEAS
Have you ever examined a specific group of samples, such as patients with a particular treatment response or cells exhibiting unique behaviors, and wondered what clinical or experimental details make them distinct? Identifying which characteristics are truly enriched or over-represented in that subset compared to the entire dataset can be surprisingly complex. This challenge is common when working with high-dimensional biomedical data.
That’s where SEAS (Statistical Enrichment Analysis of Samples) comes into play. SEAS is an interactive online tool designed for this purpose: to help you explore the clinical attributes (or “clinotypes,” such as age group, treatment status, or survival days) within your sample cohort and identify which ones are significantly over-represented compared to your overall dataset. This tool is beneficial whether you’re trying to balance case-control groups for a study or simply aiming to profile and understand a specific sample subset better. Many existing tools have struggled with smoothly integrating different data types or automatically identifying similarities between samples.
What SEAS Does
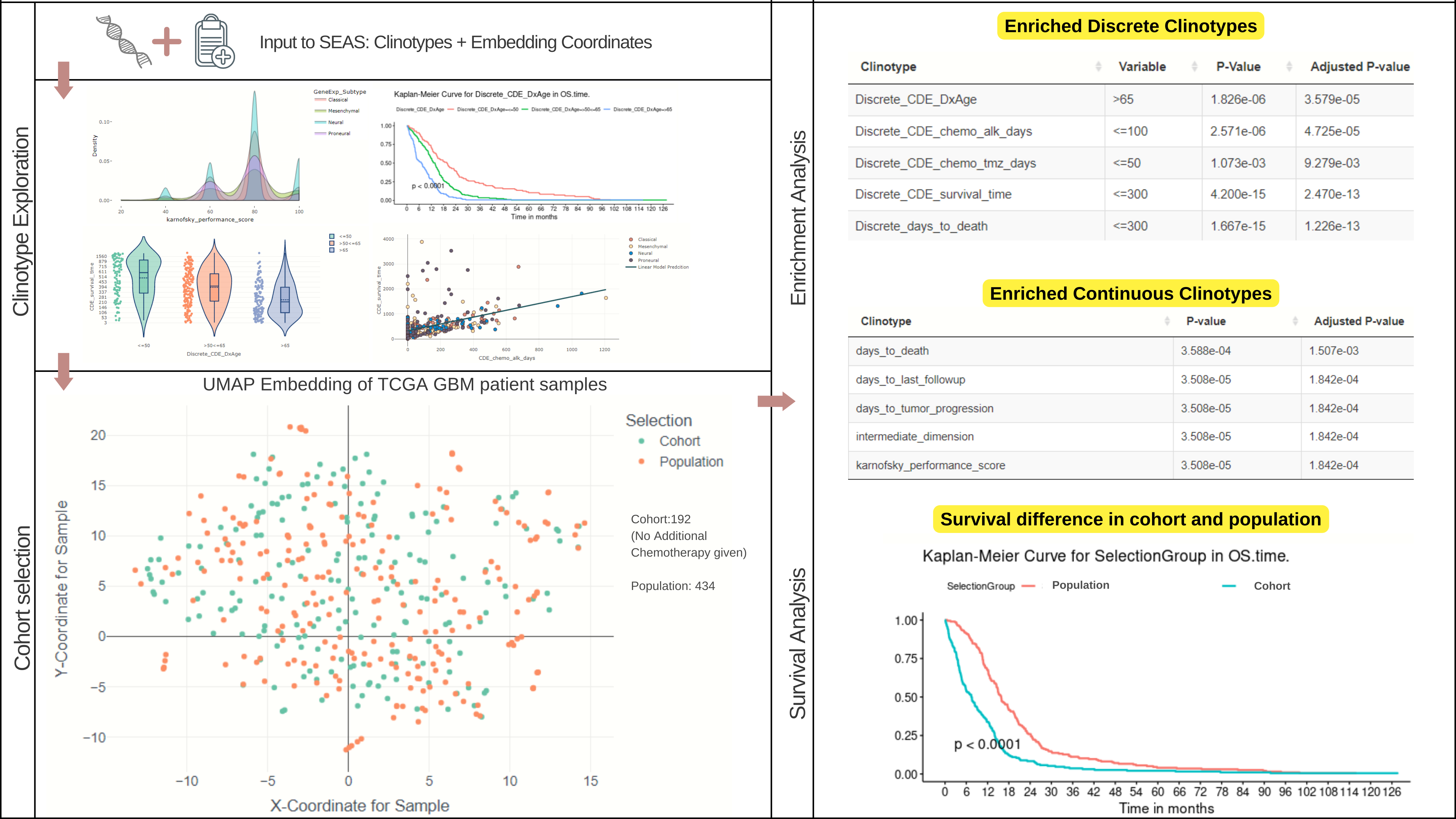
Identifies statistically enriched clinical features (clinotypes) in a user-defined sample subset.
Works with both categorical (e.g., gender, treatment type) and numerical data (e.g., age, survival time).
Provides visualizations to explore clinical features (like density plots, survival plots) and sample relationships (embedding plots).
Can help quantify and visualize similarities between patients or samples.
Supports automatic clustering to help identify meaningful subcohorts.
My Role
I developed SEAS as my undergraduate thesis project. My work involved building the tool itself and implementing various interactive components, including visualizations for exploring clinical data distribution, survival analysis, and sample embeddings, along with methods for interactive cohort selection.
How You Can Use It
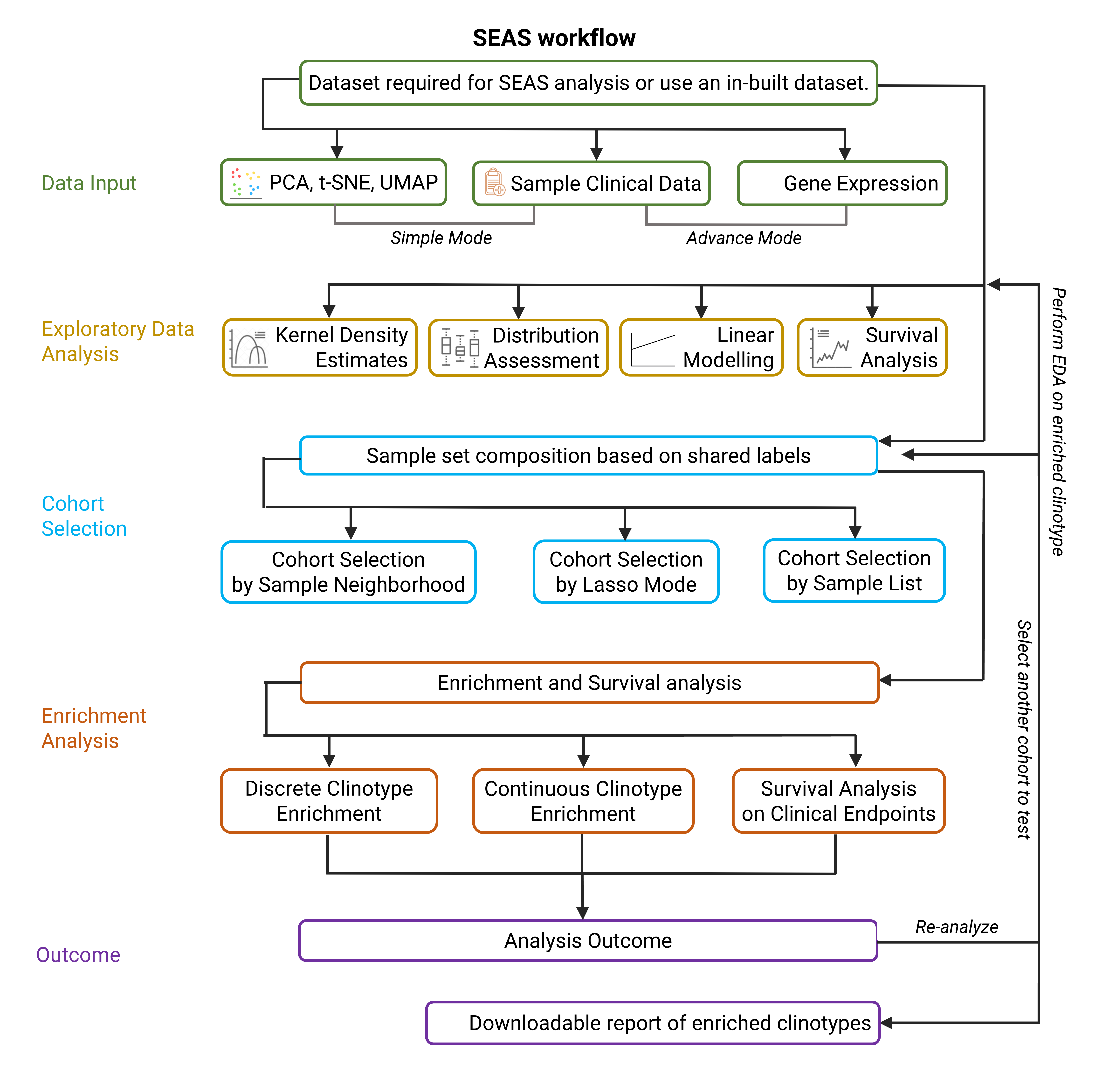
Upload your clinical metadata table.
Upload your sample embedding (generated using gene exp or similar matrices)
Explore relationships among sample clinotypes (features).
Define your sample subset of interest (you can select manually, or SEAS can help identify clusters).
SEAS performs Clinical Feature Enrichment Analysis (CFEA) and reports the features significantly enriched in your subset, complete with statistical measures.
Check it Out
Try the live SEAS tool here: https://aimed-lab.shinyapps.io/SEAS/
Find more info and documentation here: https://aimed-lab.github.io/SEAS/
References
Nguyen, T. M., Bharti, S., Yue, Z., Willey, C. D., & Chen, J. Y. (2021). Statistical Enrichment Analysis of Samples: A General-Purpose Tool to Annotate Metadata Neighborhoods of Biological Samples. Frontiers in big data, 4, 725276. https://doi.org/10.3389/fdata.2021.725276
Nguyen, T. M., Bharti, S., Yue, Z., Willey, C. D., & Chen, J. Y. (2021). Corrigendum: Statistical Enrichment Analysis of Samples: A General-Purpose Tool to Annotate Metadata Neighborhoods of Biological Samples. Frontiers in big data, 4, 804141. https://doi.org/10.3389/fdata.2021.804141
This post was drafted with assistance from various AI models to help share my project work more effectively. Please feel free to reach out if you spot any typos or have corrections!